



The Molecular Mycology Laboratory
Recent Publications
Faculty Opinions
Faculty Opinions


Faculty Opinions

Members
J C Bose National Fellow
Fellow, American Academy of Microbiology
Chair & Professor
Molecular Biology & Genetics Unit
Visiting Professor, Osaka University
PhD: Bose Institute ; Mentor: Prof. Pratima Sinha
Postdoc: UC, Santa Barbara; Mentor: Prof. John Carbon


Aswathy Narayanan
JNC Research Associate
aswathy@jncasr.ac.in

Md. Hashim Reza
DBT Research Associate
hashimreza@jncasr.ac.in

Dileep Pullepu
ICMR Research Associate
Dileep@jncasr.ac.in

Banishree Sahoo
TRC Research Associate

P V S Satya Dev
Research Associate
Rashi Aggarwal
Graduate Student
rashi@jncasr.ac.in

Kuladeep Das
Graduate Student
kuladeep@jncasr.ac.in


Srijana Dutta
Graduate Student
srijana@jncasr.ac.in

Rohit Goyal
Graduate Student
rohitgoyal@jncasr.ac.in

Shreekrishna Bhat
Graduate Student

Amrutha A S
Graduate Student
amrutha@jncasr.ac.in

Pooja Joshi
Graduate Student
pooja@jncasr.ac.in

Nidhi Ray
Graduate Student
nidhi@jncasr.ac.in


Sahana Ravi
Administrative Assistant
Nagaraj C
Lab Assistant
Alumni
PhD Students
2011 Jitendra Thakur (Assistant Professor, Emory University)
2012 Babhrubahan Roy (Ajit Joglekar Lab, University of Michigan, Ann Arbor)
2014 Sreyoshi Mitra (Tchasovnikarova Lab, Gurdon Institute, Cambridge)
2014 Gautam Chatterjee (Assistant Professor, Vivekananda University)
2014 Laxmi Shanker Rai (Christophe d’Enfert Lab, Pasteur Institute, Paris)
2017 Vikas Yadav (Joe Heitman Lab, Duke University)
2018 Neha Varshney (Arshad Desai Lab, University of California, San Diego)
2019 Lakshmi Sreekumar (Julia Cooper Lab, University of Colorado, Denver)
2020 Shreyas Sridhar (Tatsuo Fukagawa Lab, Osaka University, Osaka)
2020 Sundar Ram Sankaranarayanan (Ines Drinnenberg Lab, Curie Institute, Paris)
2020 Krishnendu Guin (Tom Misteli Lab, NCI, National Institutes of Health)
2020 Rima Singha (Jules Ene Lab, Pasteur Institute, Paris)
2021 Priya Jaitly (Robert Arkowitz Lab, iBV, Nice)
2022 Priya Brahma
2023 PVS Satya Dev (thesis submitted)
MS Students
2012 Sivani Vempati
2012 Vikas Yadav
2012 Lakshmi Sreekumar
2013 Shreyas Sridhar
2013 Sundar Ram S
2015 Priya Jaitly
2016 Jigyasa Verma
2016 Aditi Batra
2017 P V S Satya Dev
2018 Bornika Roy
2019 Kuladeep Das
2019 Rashi Aggarwal
2020 Padmalaya
2021 Srijana Dutta
2021 Rohit Goyal
2022 Harshit Arya
Postdoctoral Fellows
2008–13 Uttara Chakraborty, Assistant Professor, Manipal Institute of Regenerative Medicine, Bangalore, India
2009–10 Deepti Jain, Scientist, Lexagene, Boston, MA, USA
2011–13 Abhishek Baghela, Associate Researcher, Centre national de la recherche scientifique, Orléans, Centre-Val de Loire, France
2012–15 Arti Dumbrepatil, Science Writer, Durham, NC, USA
2016-22 Shweta Panchal, Associate Professor, Vellore Institute of Technology, TN, India
Project Fellows
2005-06 Shlini
2006–06 Shweta
2006-07 Museer Ahmed Lone
2006-08 Sreedevi Padmanabhan
2007-08 Jinal Shukla
2008–08 Rituparna Mondal
2008–11 Yogitha Thattikota
2008-10 Vikram Naik
2009–10 Ramesh Gadi
2011-13 Tanmoy Chakraborty
2012-12 Hima Bindu
2013-15 Asif Bakshi
2014-15 Parijat Bandhopadhyay
2017–17 Priti Rai
2015–17 Radha Mishra
2017–19 Abhijit Das
2017–18 Bhagya C T
2018–19 Promit Ganguly
2019-21 Tejas Patel
2020-21 Debadeep Chaudhury
2020-22 Nidhi Ray
Undergraduate Students
POBE Students
2007-08 Deepti Ramachandran
2010-11 Vishram Terse
2011-12 Hemanta Sarmah
2015–16 Ninadini Sharma
2018–19 Mohammad Abdus Shakur
IISc Undergraduate Student
2012-13 Madwesh
Summer Students
2006 Achira Roy
2006 Jinal Shukla
2006 Sreedevi Padmanabhan
2007 Anuja C
2009 Shipra Grover
2010 Shipra Grover
2013 Lavanya Balakrishnan
2014 Jyothsna Rameshkumar
2014 Devika C Namboodiri
2015 Dipasree Hajra
2015 Bhavya Revathy
2016 Upasana Basu
2016 Chitra Joshi
2017 Kajal Sandhu
2018 Arkaprabha Adhikary
2019 Deepak M. Kushalani
Short-term Visitors
Aditya Singh Pratihar (Purvanchal University, India)
Jonathan Gomez Raja (Universidad de Extremadura, Spain)
Priya (IIT-Bombay, India)
Aakansha (IIT-Bombay)
Sabyasachi Sutradhar (IACS, India)
Song Hyungnam (KRIBB, Korea)
Visiting Scientists
2008 Dharanidhar Dubey (Professor, Purvanchal University, India)
2009 Dharanidhar Dubey (Professor, Purvanchal University, India)
2010 Indrani Bose (Associate Professor, Western Carolina University, USA)
2013 Euijeon Woo (Professor, KRIBB, Korea)
2018 Bharati Prakash (Assistant Professor, University College-Mangalore, India)
Publications






-
Coelho MA, Ianiri G, David-Palma M, Theelen B, Goyal R, Narayanan A, Lorch JM, Sanyal K, Boekhout T, Heitman J (2023) Frequent transitions in mating-type locus chromosomal organization in Malassezia and early steps in sexual reproduction. PNAS PDF
-
Singha R, Aggarwal R, Sanyal K* (2023) Negative regulation of biofilm development by the CUG-Ser1 clade-specific histone H3 variant is dependent on the canonical histone chaperone CAF1 complex in Candida albicans. Molecular Microbiology (in press)
-
Narayanan A^, Kumar P^, Chauhan A, Kumar M, Yadav K, Banerjee A, Sharma RD, Rudramurthy SM, Chakrabarti A, Sanyal K*, Prasad R* (2022) Directed evolution detects supernumerary centric chromosomes conferring resistance to azoles in Candida auris. mBio PDF
-
Jaitly P, Legrand M, Das A, Patel T, Chauvel M, Maufrais C, d’Enfert C*, Sanyal K* (2022) A phylogenetically-restricted essential cell cycle progression factor in the human pathogen Candida albicans. Nature Communications PDF Recommended by F1000
-
Narayanan A*, Selvakumar P, Siddharthan R, Sanyal K* (2022) ClaID: A rapid method of clade-level identification of the multidrug-resistant human fungal pathogen Candida auris. Microbiology Spectrum PDF
-
Panchal S*, Sanyal K* (2022) Loss of nucleosome assembly protein 1 affects growth and appressorium structure in blast fungus Magnaporthe oryzae. microPublication Biology PDF
-
Rudramurthy S, Kashyap N, Bhattacharya S, Soman R, Shankarnarayan S, Chavan D, Singh S, Das P, Kaur H, Ghosh A, Prasad S, Sanyal K, and Chakrabarti A (2022) Impact of FKS1 genotype on echinocandin in-vitro susceptibility in Candida auris and in-vivo response in a murine model of infection. Antimicrobial Agents and Chemotherapy PDF
-
Mehta G, Sanyal K, Abhishek S, Rajakumara E, Ghosh SK (2022) Minichromosome maintenance proteins in eukaryotic chromosome segregation. BioEssays PDF
-
Padmanabhan S, Sanyal K*, Dubey DD* (2021) Identification and in silico analysis of the origin recognition complex in the human fungal pathogen Candida albicans. microPublication Biology PDF
-
Sane A, Sridhar S, Sanyal K, Ghosh SK (2021) Shugoshin ensures maintenance of the spindle assembly checkpoint response and efficient spindle disassembly. Molecular Microbiology PDF
-
Chatterjee S, Som S, Varshney N, Satyadev PVS, Sanyal K, Paul R (2021) Mechanics of microtubule organizing center clustering and spindle positioning in budding yeast Cryptococcus neoformans. Physical Review E PDF
-
Reza H*, Patkar R*, Sanyal K* (2021) Vacuolar transporter Mnr2 safeguards organellar integrity in aged cells. Molecular Microbiology PDF
-
Narayanan A, Vadnala RN, Ganguly P, Selvakumar P, Rudramurthy SM, Prasad R, Chakrabarti A, Siddharthan R, Sanyal K* (2021) Functional and comparative analysis of centromeres reveals clade-specific rearrangements in Candida auris and a chromosome number change in related species. mBio. PDF Commentary I Stable Positions of Epigenetically Inherited Centromeres in the Emerging Fungal Pathogen Candida auris and Its Relatives. mBio. PDF Commentary I The curious case of nonrepetitive centromeric DNA sequences in Candida auris and related species . mBio. PDF
-
Sreekumar L, Kumari K, Guin K, Bakshi A, Varshney N, Thimmappa BC, Narlikar L, Padinhateeri R, Siddharthan R, Sanyal K* (2021) Orc4 spatiotemporally stabilizes centromeric chromatin. Genome Research. PDF
-
Sridhar S, Hori T, Nakagawa R, Fukagawa T*, Sanyal K* (2021) Bridgin connects the outer kinetochore to centromeric chromatin. Nature Communications. PDF Recommended by F1000
-
Guin K^*, Sreekumar L^*, Sanyal K* (2020) Implications of the evolutionary trajectory of centromeres in the fungal kingdom. Annual Review of Microbiology. PDF; Invited Review
-
Guin K, Chen Y, Mishra R, Siti R, Thimmappa BC, O'Brien C, Butler G, Sanyal A*, Sanyal K* (2020) Spatial inter-centromeric interactions facilitated the emergence of evolutionary new centromeres. eLIFE. PDF Recommended by F1000
-
Fang YF, Coelho MA, Sue H, Schotanus K, Thimmappa BC, ...., Sanyal K, Dong S, Nowrousian M, Heitman J (2020) Long transposon-rich centromeres in an oomycete reveal divergence of centromere features in Stramenopila-Alveolata-Rhizaria lineages. PLOS Genetics. PDF
-
Sankaranarayanan SR, Ianiri G, Coelho M, Reza H, Thimmappa BC, Ganguly P, Vadnala RN, Sun S, Siddharthan R, Tellgren-Roth C, Dawson TL, Heitman J*, Sanyal K* (2020) Loss of centromere function drives karyotype evolution in closely related Malassezia species. eLIFE. PDF eLife digest The yeast that re-organized its centromeres
-
Navarro-Mendoza M, Perez-Arques C, Panchal S, Nicolas FE, Mondo SJ, Ganguly P, Pangilinan J, Grigoriev IV, Heitman J*, Sanyal K* and Garre V* (2019) Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Current Biology. PDF Recommended by F1000 ; Commentary I Evolution: A mosaic-type centromere in an early diverging fungus. Current Biology. PDF
-
Rai LS, Singha R, Sanchez H, Chakraborty T, Chand B, Bachellier-Bassi S, Chowdhury S, d’Enfert C, Andes DR, Sanyal K* (2019) The Candida albicans biofilm gene circuit modulated at the chromatin level by a recent molecular histone innovation, PLOS Biology. PDF Recommended by F1000
-
Yadav V^, Yang F^, Reza H, Liu S, Valent B, Sanyal K*, Naqvi NI* (2019) Cellular dynamics and genomic identity of centromeres in cereal blast fungus, mBIO. PDF
-
Prasad P, Sanyal K, Ghosh SK (2019) Sth1, the key subunit of the RSC chromatin remodeling complex, is essential in maintaining chromosomal integrity and mediating high fidelity chromosome segregation in the human fungal pathogen Candida albicans, Frontiers in Microbiology. PDF
-
Varshney N*, Sanyal K* (2019) Nuclear migration in budding yeasts: position before division, Current Genetics. PDF; Invited Review
-
Sreekumar L, Jaitly P, Chen Y, Thimmappa BC, Sanyal A, Sanyal K* (2019) Cis and trans chromosomal interactions define pericentric boundaries in the absence of conventional heterochromatin, Genetics. PDF
-
Suneet K^, Sridhar S^, Agiwal P, Sridhar MS, Sanyal K, Jain S (2019) Magnetic hyperthermia adjunctive therapy for fungi: in vitro studies against Candida albicans, International Journal of Hyperthermia. PDF
-
Varshney N, Sanyal K* (2019) Aurora kinase Ipl1 facilitates bilobed distribution of clustered kinetochores to ensure error‐free chromosome segregation in Candida albicans, Molecular Microbiology. PDF
-
Legrand M, Jaitly P, Feri A, d’Enfert C*, Sanyal K* (2019) Candida albicans: an emerging yeast model to study eukaryotic genome plasticity, Trends in Genetics. PDF; Invited Review
-
Varshney N, Som S^, Chatterjee S^, Sridhar S, Bhattacharyya D, Paul R*, Sanyal K* (2019) Spatio-temporal regulation of nuclear division by Aurora B kinase Ipl1 in Cryptococcus neoformans, PLOS Genetics. PDF
-
Kakade P, Mahadik K, Balaji KN, Sanyal K, Nagaraja V (2019) Two negative regulators of biofilm development exhibit functional divergence in conferring virulence potential to Candida albicans, FEMS Yeast Research. PDF
-
Hoque J, Yadav V, Prakash RG, Sanyal K, Haldar J (2018) Dual-Function Polymer–Silver Nanocomposites for Rapid Killing of Microbes and Inhibiting Biofilms, ACS Biomaterials Science & Engineering. PDF
-
Yadav V, Sreekumar L, Guin K, Sanyal K* (2018) Five pillars of centromeric chromatin in fungal pathogens, PLOS Pathogens. PDF Invited Review
-
Yadav V*, Sanyal K* (2018) Sad1 spatiotemporally regulates kinetochore clustering to ensure high-fidelity chromosome segregation in the human fungal pathogen Cryptococcus neoformans, mSphere. PDF
-
Yadav V, Sun S, Billmyre RB, Thimmappa BC, Shea T, Lintner R, Bakkeren G, Cuomo CA, Heitman J, Sanyal K* (2018) RNAi is a critical determinant of centromere evolution in closely related fungi, PNAS. PDF
-
Rai LS, Singha R, Brahma P, Sanyal K*. (2018) Epigenetic determinants of phenotypic plasticity in Candida albicans, Fungal Biology Reviews. PDF; Invited Review
-
Sun S, Yadav V, Billmyre RB, Cuomo CA, Nowrousian M, Wang L, Souciet JL, Boekhout T, Porcel B, Wincker P, Granek JA, Sanyal K, Heitman J. (2017) Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination, PLOS Biology. PDF
-
Altamirano S, Fang D, Simmons C, Sridhar S, Wu P, Sanyal K, Kozubowski L. (2017) Fluconazole-induced ploidy change in Cryptococcus neoformans results from the uncoupling of cell growth and nuclear division, mSphere. PDF; Recommended by F1000
-
Ghosh C, Yadav V, Younis W, Mohammad H, Hegazy YA, Seleem MN, Sanyal K, Haldar J. (2017) Aryl-alkyl-lysines: membrane-active fungicides that act against biofilms of Candida albicans, ACS Infectious Diseases. PDF
-
Zhu Y, Engström PG, Tellgren-Roth C, Baudo CD, Kennell JC, Sun S, Billmyre RB, Schröder MS, Sankaranarayanan SR, Sanyal K, Andersson A, Holm T, Sigurgeirsson B. (2017) Proteogenomics produces comprehensive and highly accurate protein-coding gene annotation in a complete genome assembly of Malassezia sympodialis. Nucleic Acids Research. PDF
-
Hoque J, Adhikary U, Yadav V, Samaddar S, Konai MM, Prakash RG, Paramanandham K, Shome BR, Sanyal K, Haldar J. (2016) Chitosan derivatives active against multidrug-resistant bacteria and pathogenic fungi: in vivo evaluation as topical antimicrobials, Molecular Pharmaceutics. PDF
-
Datta A^, Yadav V^, Ghosh A, Choi J, Bhattacharyya D, Kar RK, Ilyas H, Dutta A, An E, Mukhopadhyay J, Sanyal K, Lee D, Ramamoorthy, A Bhunia A (2016) Mode of action of a designed antimicrobial peptide: high potency against Cryptococcus neoformans, Biophysical Journal. PDF
-
Kakade P, Sadhale P, Sanyal K, Nagaraja V. (2016) ZCF32, a fungus-specific Zn (II) 2 Cys6 transcription factor, is a repressor of the biofilm development in the human pathogen Candida albicans, Scientific Reports. PDF
-
Chatterjee G, Sankaranarayanan SR, Guin K, Thattikota Y, Padmanabhan S, Siddharthan R, Sanyal K* (2016) Repeat-associated fission yeast-like regional centromeres in the ascomycetous budding yeast Candida tropicalis, PLOS Genetics. PDF
-
Mitra S, Rai LS, Chatterjee G, Sanyal K* (2016) Chromatin immunoprecipitation (ChIP) assay in Candida albicans, Methods in Molecular Biology. PDF
-
Suryanarayanan TS, Gopal V, Sahal D and Sanyal K (2015) Establishing a national fungal genetic resource to enhance the bioeconomy, Current Science. PDF
-
Sutradhar S^ Yadav V^, Sridhar S^, Sreekumar L, Bhattacharyya D, Ghosh SK, Paul R* and Sanyal K* (2015) A comprehensive model to predict mitotic division in budding yeasts, Molecular Biology of the Cell. PDF
-
KT Nishant* and Sanyal K* (2015) The good, the bad and the ugly: How to protect genome stability from potential threats, Bioessays. PDF Meeting Highlights
-
Varshney N^, Schakel A^, Singha R, Chakraborty T, Wijlick LV, Ernst JF* and Sanyal K* (2015), A surprising role for the Sch9 protein kinase in chromosome segregation in Candida albicans, Genetics. PDF
-
Hoque J, Akkapeddi P, Yadav V, Manjunath GB, Uppu DSSM, Konai MM, Yarlagadda V, Sanyal K and Haldar J (2015) Broad-spectrum antibacterial and antifungal polymeric paint materials: synthesis, structure-activity relationship and membrane-active mode of action, ACS Appl. Mater. Interfaces. PDF
-
Mitra S, Gomez-Raja J, Larriba G, Dubey DD and Sanyal K* (2014) Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans, PLOS Genetics. PDF
-
Janbon G, Ormerod KL, Paulet D, Byrnes III EJ, Yadav V, Chatterjee G, (+41 authors), Sanyal K, Heitman J, Fraser JA, Cuomo CA and Dietrich FS (2014) Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation, PLOS Genetics. PDF; Recommended by F1000
-
Kozubowski L^, Yadav V^, Chatterjee G, Sridhar S, Yamaguchi M, Kawamoto S, Bose I, Heitman J and Sanyal K* (2013) Ordered kinetochore assembly in the human fungal pathogen basidiomycetous yeast Cryptococcus neoformans, mBIO. PDF
-
Chakraborty U, Mohamed A, Kakade P, Mugasimangalam R, Sadhale PP and Sanyal K* (2013) A stable hybrid containing haploid genomes of two obligate diploid Candida species, Eukaryotic Cell. PDF
-
Thakur J and Sanyal K* (2013) Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans, Genome Research. PDF
-
Roy B, Varshney N, Yadav V and Sanyal K* (2013) The process of kinetochore assembly in yeasts, FEMS Microbiology Letters. PDF Invited Review
-
Thakur J and Sanyal K* (2012) A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans, PLOS Genetics. PDF
-
Sanyal K* (2012) How do microbial pathogens make CENs?, PLOS Pathogens. PDF Invited Review
-
Laha S, Das SP, Hajra S, Sanyal K and Sinha P (2011) Functional characterization of the Saccharomyces cerevisiae protein Chl1 reveals the role of sister chromatid cohesion in the maintenance of spindle length during S-phase arrest, BMC Genetics. PDF
-
Roy B and Sanyal K* (2011) Diversity in Requirement of Genetic and Epigenetic Factors for Centromere Function in Fungi, Eukaryotic Cell. PDF Invited Review
-
Thakur J and Sanyal K* (2011) The essentiality of the fungus specific Dam1 complex is correlated with one kinetochore one microtubule interaction present throughout the cell cycle, independent of the nature of a centromere, Eukaryotic Cell. PDF
-
Roy B, Burrack LS, Lone MA, Berman J and Sanyal K* (2011) CaMtw1, a member of the evolutionarily conserved Mis12 kinetochore protein family, is required for efficient inner kinetochore assembly in the pathogenic yeast Candida albicans, Molecular Microbiology. PDF
-
Padmanabhan S^, Thakur J^, Siddharthan R and Sanyal K* (2008) Rapid evolution of Cse4p-rich centromeric DNA sequences in closely-related pathogenic yeasts, Candida albicans and Candida dubliniensis, PNAS. PDF Recommended by F1000
-
Baum M^, Sanyal K^, Mishra PK, Thaler N and Carbon J (2006) Formation of functional centromeric chromatin is specified epigenetically in Candida albicans, PNAS. PDF; Recommended by F1000
-
Sanyal K, Baum M and Carbon J (2004) Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique, PNAS. PDF
-
Sanyal K and Carbon J (2002) The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans in a centromere protein essential for chromosomes transmission, PNAS. PDF
-
Ghosh SK, Poddar A, Hajra S, Sanyal K and Sinha P (2001) The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation, Molecular Genetics and Genomics. PDF
-
Sanyal K, Ghosh SK and Sinha P (1998) The MCM16 gene of the yeast Saccharomyces cerevisiae is required for chromosome segregation, Molecular and General Genetics. PDF ^ Equally contributed; *Corresponding author
US Patent
Sanyal K, Padmanabhan S, Thakur J (2014) Polynucleotide sequences of Candida dubliniensis and probes for its detection.
Funding





Student Awards
2021
-
Sundar Ram S, EMBO Long-term postdoctoral fellowship
-
Neha Varshney, NASI Young Scientist Award
2020
-
Krishnendu Guin, CNR Rao Medal for Best Thesis in Biological Sciences
2019
-
Vikas Yadav, INSA Medal for Young Scientists
-
Vikas Yadav, NASI Young Scientist Award
-
Neha Varshney, Research Associate fellowship from CSIR
-
Shreyas Sridhar, EMBO travel grant to attend “ Chromosome segregation and Aneuploidy meeting” at Cascais, Portugal.
-
Md. Hashim Reza, travel fellowship from SERB to attend “30th Fungal Genetics Conference” at Pacific Grove, California, USA
-
Md. Hashim Reza, DBT-CTEP travel Grant to attend “The 8th International Rice Blast Conference” held at Chengdu, China
2018
-
Jitendra Thakur, INSA Medal for Young Scientists
-
Neha Varshney, PLOS Genetics Best Poster Award in Chromosome Stability 2018 in Bangalore
-
Krishnendu Guin, PLOS Genetics Best Poster Award in Chromosome Stability 2018 in Bangalore
-
Lakshmi Sreekumar, PLOS Genetics Best Poster Award in Chromosome Stability 2018 in Bangalore.
Gallery

Kaustuv Sanyal with Louise Clarke and John Carbon

Jinal, Sreedevi, Achira (summer trainees in 2006)

Lab lunch at Byg Brewski (2021)

Kaustuv Sanyal with Louise Clarke and John Carbon
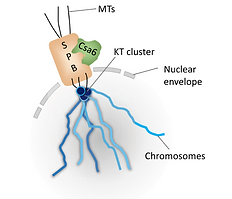
Organization of SPB (Alp6-mCherry), kinetochores (GFP-CenpA) and microtubules (GFP-TubA) during mitosis in M. oryzae
Live‐cell time‐lapse imaging of CaMtw1–GFP in hyphal cells of Candida albicans.
Live‐cell time‐lapse imaging of CaMtw1–GFP in yeast cells of Candida albicans.
Contact Us
Kaustuv Sanyal, PhD
Molecular Mycology Laboratory
Molecular Biology and Genetics Unit
Jawaharlal Nehru Centre for Advanced Scientific Research
Rachenahalli Lake Road, Jakkur,
Bengaluru, Karnataka-560064
India
Telephone: +91 80-2208-2898 (lab I), +91 80-2208-2704 (lab II), +91 80 2208 2878 (office)
email: sanyal@jncasr.ac.in
Nuclear migration in the wild-type and Ipl1-depleted cells of C. neoformans. The nucleus is visualized by GFP-tagged histone H4. Images were acquired every 60 s with confocal microscopy.
Movies
MTOC clustering and nuclear migration in the wild-type and ipl1 mutant with the effect of biased cortical interaction of C. neoformans
Kinetochores decluster momentarily but arrange randomly on the spindle axis before sister kinetochore separation during anaphase in M. oryzae.


